Assessment of Promoter hypermethylation of APC and BRCA1 in endometrial cancer.
Shivanandappa N.1, N Swamy S.2, Kumar S S.3, Sheshadri S.4, Venkateshaiah Reddihalli P.5, Gawari R.6*
DOI: https://doi.org/10.17511/ijmrr.2021.i04.06
1 Nagaratna Shivanandappa, Research Scholar, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
2 Shalini N Swamy, Research Scholar, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
3 Sandeep Kumar S, Senior Research Fellow, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
4 Suma Sheshadri, Associate Professor, Department of Pathology, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
5 Pallavi Venkateshaiah Reddihalli, Professor, Department of Gynaecology, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
6* Ramesh Gawari, Professor, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
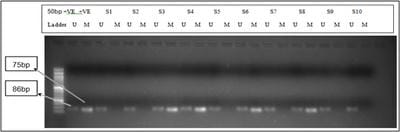
Introduction: Endometrial cancer is one of the most common cancers in women worldwide. The underlying cause of endometrial tumorigenesis remains elusive. Several genetic and epigenetic alterations are known to be involved in the carcinogenesis of endometrial carcinoma. One important and early epigenetic alteration that is attributed to endometrial carcinoma is the aberrant promoter hypermethylation of gene promoters. In this study, we have assessed the aberrant promoter hypermethylation of APC and BRCA1 in 78 endometrial cancer samples. Methods: Histologically confirmed tumour tissue samples were obtained post-surgery and DNA was extracted. The DNA was subjected to sodium bisulfite conversion and used as a template for a polymerase chain reaction. The PCR was performed using a nested PCR followed by methylation specific PCR. Results: A 33.33% and 46.15% methylation frequency was observed for APC and BRCA1 genes respectively. A higher percentage of methylation was observed in stage IV for APC (66.66%) and in stage II for BRCA1 (88.88%). Conclusion: Aberrant promoter hypermethylation is an early event in endometrial carcinoma and can serve as a useful molecular marker for diagnosis and prognosis of the disease along with existing screening modalities.
Keywords: Hypermethylation, Endometrial cancer, APC, BRCA1
| Corresponding Author | How to Cite this Article | To Browse |
|---|---|---|
| , Professor, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India. Email: |
Shivanandappa N, Swamy SN, Kumar SS, Sheshadri S, Reddihalli PV, Gawari R. Assessment of Promoter hypermethylation of APC and BRCA1 in endometrial cancer.. Int J Med Res Rev. 2021;9(4):241-248. Available From https://ijmrr.medresearch.in/index.php/ijmrr/article/view/1306 |


 ©
©