Aberrant promoter hypermethylation of RAR-β in endometrial carcinoma- an Indian study.
Shivanandappa N.1, N Swamy S.2, Kumar S S.3, Sheshadri S.4, Reddihalli Pallavi V.5, Gawari R.6*
DOI: https://doi.org/10.17511/ijmrr.2021.i03.05
1 Nagaratna Shivanandappa, Research Scholar, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
2 Shalini N Swamy, Research Scholar, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
3 Sandeep Kumar S, Senior Research Fellow, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
4 Suma Sheshadri, Associate Professor, Department of Pathology, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
5 Venkateshaiah Reddihalli Pallavi, Professor, Department of Gynaecology, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
6* Ramesh Gawari, Professor, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India.
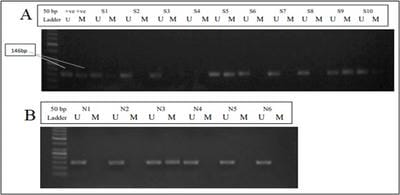
Endometrial cancer is the seventh most common cancer in women worldwide with an age-standardized rate of 8.4 per 100,000 women. Epigenetic alterations such as promoter hypermethylation of TSGs are known to be early events in carcinogenesis. The aim of the present study, we assessed the aberrant promoter hypermethylation pattern of RAR-β in 78 endometrial cancer samples. Methods: DNA was isolated from endometrial carcinoma samples and normal tissues and aberrant promoter hypermethylation was assessed using nested and methylation-specific PCR. The Chi-square test was used for statistical analysis and a p-value<0.05 was considered to be statistically significant. Results: 40 of the 78 (51.28%) endometrial carcinoma samples showed aberrant hypermethylation of the RAR-β gene. Methylation status in each histological subtype, grade and stage of the disease was also assessed. Conclusion: Aberrant hypermethylation is an important early epigenetic alteration that occurs during tumorigenesis. The Data shown here reports that promoter hypermethylation of RAR-β occurs in endometrial carcinoma and therefore could be used as a potential marker for early diagnosis and prognosis of the disease.
Keywords: DNA Hypermethylation, RAR-β, Endometrial cancer, epigenetic alterations, DNA hypermethylation, Methylation-specific PCR
| Corresponding Author | How to Cite this Article | To Browse |
|---|---|---|
| , Professor, Department of Biochemistry, Kidwai Memorial Institute of Oncology, Bangalore, Karnataka, India. Email: |
Shivanandappa N, Swamy SN, Kumar S S, Sheshadri S, Venkateshaiah Reddihalli Pallavi, Ramesh Gawari, Aberrant promoter hypermethylation of RAR-β in endometrial carcinoma- an Indian study.. Int J Med Res Rev. 2020;9(3):160-166. Available From https://ijmrr.medresearch.in/index.php/ijmrr/article/view/1294 |


 ©
©